Convolutional variational autoencoder with PyMC3 and Keras¶
In this document, I will show how autoencoding variational Bayes (AEVB) works in PyMC3’s automatic differentiation variational inference (ADVI). The example here is borrowed from Keras example, where convolutional variational autoencoder is applied to the MNIST dataset. The network architecture of the encoder and decoder are completely same. However, PyMC3 allows us to define the probabilistic model, which combines the encoder and decoder, in the way by which other general probabilistic models (e.g., generalized linear models), rather than directly implementing of Monte Carlo sampling and the loss function as done in the Keras example. Thus I think the framework of AEVB in PyMC3 can be extended to more complex models such as latent dirichlet allocation.
- Notebook Written by Taku Yoshioka (c) 2016
For using Keras with PyMC3, we need to choose Theano as the backend of Keras.
Install required packages, including pymc3, if it is not already available:
In [1]:
#!pip install --upgrade keras
#!pip install theano==0.8.0
In [2]:
#!pip install --upgrade pymc3
In [3]:
#!conda install -y mkl-service
In [1]:
%autosave 0
%matplotlib inline
import sys, os
%env KERAS_BACKEND=theano
%env THEANO_FLAGS=device=cuda3,floatX=float32,optimizer=fast_run
from collections import OrderedDict
from keras.layers import InputLayer, BatchNormalization, Dense, Conv2D, Deconv2D, Activation, Flatten, Reshape
import numpy as np
import pymc3 as pm
from theano import shared, config, function, clone, pp
import theano.tensor as tt
import keras
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import seaborn as sns
from keras import backend as K
K.set_image_dim_ordering('th')
Autosave disabled
env: KERAS_BACKEND=theano
env: THEANO_FLAGS=device=cuda3,floatX=float32,optimizer=fast_run
Using Theano backend.
Using cuDNN version 5105 on context None
Mapped name None to device cuda3: Tesla K40m (0000:84:00.0)
In [2]:
import pymc3, theano
print(pymc3.__version__)
print(theano.__version__)
print(keras.__version__)
3.1rc3
0.9.0.dev-f4bb35d6264213d023706b7baa065a60c6416149
2.0.4
Load images¶
MNIST dataset can be obtained by scikit-learn API or from Keras datasets. The dataset contains images of digits.
In [3]:
# from sklearn.datasets import fetch_mldata
# mnist = fetch_mldata('MNIST original')
# print(mnist.keys())
from keras.datasets import mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
data = pm.floatX(x_train.reshape(-1, 1, 28, 28))
data /= np.max(data)
Downloading data from https://s3.amazonaws.com/img-datasets/mnist.npz
Use Keras¶
We define a utility function to get parameters from Keras models. Since we have set the backend to Theano, parameter objects are obtained as shared variables of Theano.
In the code, ‘updates’ are expected to include update objects
(dictionary of pairs of shared variables and update equation) of scaling
parameters of batch normalization. While not using batch normalization
in this example, if we want to use it, we need to pass these update
objects as an argument of theano.function() inside the PyMC3 ADVI
function. The current version of PyMC3 does not support it, it is easy
to modify (I want to send PR in future).
The learning phase below is used for Keras to known the learning phase, training or test. This information is important also for batch normalization.
In [5]:
from keras.models import Sequential
from keras.layers import Dense, BatchNormalization
def get_params(model):
"""Get parameters and updates from Keras model
"""
shared_in_updates = list()
params = list()
updates = dict()
for l in model.layers:
attrs = dir(l)
# Updates
if 'updates' in attrs:
updates.update(l.updates)
shared_in_updates += [e[0] for e in l.updates]
# Shared variables
for attr_str in attrs:
attr = getattr(l, attr_str)
if isinstance(attr, tt.compile.SharedVariable):
if attr is not model.get_input_at(0):
params.append(attr)
return list(set(params) - set(shared_in_updates)), updates
# This code is required when using BatchNormalization layer
keras.backend.theano_backend._LEARNING_PHASE = \
shared(np.uint8(1), name='keras_learning_phase')
Encoder and decoder¶
First, we define the convolutional neural network for encoder using Keras API. This function returns a CNN model given the shared variable representing observations (images of digits), the dimension of latent space, and the parameters of the model architecture.
In [6]:
def cnn_enc(xs, latent_dim, nb_filters=64, nb_conv=3, intermediate_dim=128):
"""Returns a CNN model of Keras.
Parameters
----------
xs : theano.TensorVariable
Input tensor.
latent_dim : int
Dimension of latent vector.
"""
input_layer = InputLayer(input_tensor=xs,
batch_input_shape=xs.tag.test_value.shape)
model = Sequential()
model.add(input_layer)
cp1 = {'padding': 'same', 'activation': 'relu'}
cp2 = {'padding': 'same', 'activation': 'relu', 'strides': (2, 2)}
cp3 = {'padding': 'same', 'activation': 'relu', 'strides': (1, 1)}
cp4 = cp3
model.add(Conv2D(1, (2, 2), **cp1))
model.add(Conv2D(nb_filters, (2, 2), **cp2))
model.add(Conv2D(nb_filters, (nb_conv, nb_conv), **cp3))
model.add(Conv2D(nb_filters, (nb_conv, nb_conv), **cp4))
model.add(Flatten())
model.add(Dense(intermediate_dim, activation='relu'))
model.add(Dense(2 * latent_dim))
return model
Then we define a utility class for encoders. This class does not depend
on the architecture of the encoder except for input shape (tensor4
for images), so we can use this class for various encoding networks.
In [7]:
class Encoder:
"""Encode observed images to variational parameters (mean/std of Gaussian).
Parameters
----------
xs : theano.tensor.sharedvar.TensorSharedVariable
Placeholder of input images.
dim_hidden : int
The number of hidden variables.
net : Function
Returns
"""
def __init__(self, xs, dim_hidden, net):
model = net(xs, dim_hidden)
self.model = model
self.xs = xs
self.out = model.get_output_at(-1)
self.means = self.out[:, :dim_hidden]
self.rhos = self.out[:, dim_hidden:]
self.params, self.updates = get_params(model)
self.enc_func = None
self.dim_hidden = dim_hidden
def _get_enc_func(self):
if self.enc_func is None:
xs = tt.tensor4()
means = clone(self.means, {self.xs: xs})
rhos = clone(self.rhos, {self.xs: xs})
self.enc_func = function([xs], [means, rhos])
return self.enc_func
def encode(self, xs):
# Used in test phase
keras.backend.theano_backend._LEARNING_PHASE.set_value(np.uint8(0))
enc_func = self._get_enc_func()
means, _ = enc_func(xs)
return means
def draw_samples(self, xs, n_samples=1):
"""Draw samples of hidden variables based on variational parameters encoded.
Parameters
----------
xs : numpy.ndarray, shape=(n_images, 1, height, width)
Images.
"""
# Used in test phase
keras.backend.theano_backend._LEARNING_PHASE.set_value(np.uint8(0))
enc_func = self._get_enc_func()
means, rhos = enc_func(xs)
means = np.repeat(means, n_samples, axis=0)
rhos = np.repeat(rhos, n_samples, axis=0)
ns = np.random.randn(len(xs) * n_samples, self.dim_hidden)
zs = means + pm.distributions.dist_math.rho2sd(rhos) * ns
return zs
In a similar way, we define the decoding network and a utility class for decoders.
In [8]:
def cnn_dec(zs, nb_filters=64, nb_conv=3, output_shape=(1, 28, 28)):
"""Returns a CNN model of Keras.
Parameters
----------
zs : theano.tensor.var.TensorVariable
Input tensor.
"""
minibatch_size, dim_hidden = zs.tag.test_value.shape
input_layer = InputLayer(input_tensor=zs,
batch_input_shape=zs.tag.test_value.shape)
model = Sequential()
model.add(input_layer)
model.add(Dense(dim_hidden, activation='relu'))
model.add(Dense(nb_filters * 14 * 14, activation='relu'))
cp1 = {'padding': 'same', 'activation': 'relu', 'strides': (1, 1)}
cp2 = cp1
cp3 = {'padding': 'valid', 'activation': 'relu', 'strides': (2, 2)}
cp4 = {'padding': 'same', 'activation': 'sigmoid'}
output_shape_ = (minibatch_size, nb_filters, 14, 14)
model.add(Reshape(output_shape_[1:]))
model.add(Deconv2D(nb_filters, (nb_conv, nb_conv), data_format='channels_first', **cp1))
model.add(Deconv2D(nb_filters, (nb_conv, nb_conv), data_format='channels_first', **cp2))
output_shape_ = (minibatch_size, nb_filters, 29, 29)
model.add(Deconv2D(nb_filters, (2, 2), data_format='channels_first', **cp3))
model.add(Conv2D(1, (2, 2), **cp4))
return model
In [9]:
class Decoder:
"""Decode hidden variables to images.
Parameters
----------
zs : Theano tensor
Hidden variables.
"""
def __init__(self, zs, net):
model = net(zs)
self.model = model
self.zs = zs
self.out = model.get_output_at(-1)
self.params, self.updates = get_params(model)
self.dec_func = None
def _get_dec_func(self):
if self.dec_func is None:
zs = tt.matrix()
xs = clone(self.out, {self.zs: zs})
self.dec_func = function([zs], xs)
return self.dec_func
def decode(self, zs):
"""Decode hidden variables to images.
An image consists of the mean parameters of the observation noise.
Parameters
----------
zs : numpy.ndarray, shape=(n_samples, dim_hidden)
Hidden variables.
"""
# Used in test phase
keras.backend.theano_backend._LEARNING_PHASE.set_value(np.uint8(0))
return self._get_dec_func()(zs)
Generative model¶
We can construct the generative model with PyMC3 API and the functions and classes defined above. We set the size of mini-batches to 100 and the dimension of the latent space to 2 for visualization.
In [10]:
# Constants
minibatch_size = 100
dim_hidden = 2
A placeholder of images is required to which mini-batches of images will
be placed in the ADVI inference. It is also the input to the encoder. In
the below, enc.model is a Keras model of the encoder network, thus
we can check the model architecture using the method summary().
In [43]:
# Placeholder of images
xs_t = tt.tensor4(name='xs_t')
xs_t.tag.test_value = np.zeros((minibatch_size, 1, 28, 28)).astype('float32')
# Encoder
enc = Encoder(xs_t, dim_hidden, net=cnn_enc)
enc.model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_10 (InputLayer) (100, 1, 28, 28) 0
_________________________________________________________________
conv2d_19 (Conv2D) (100, 1, 28, 28) 5
_________________________________________________________________
conv2d_20 (Conv2D) (100, 64, 14, 14) 320
_________________________________________________________________
conv2d_21 (Conv2D) (100, 64, 14, 14) 36928
_________________________________________________________________
conv2d_22 (Conv2D) (100, 64, 14, 14) 36928
_________________________________________________________________
flatten_4 (Flatten) (100, 12544) 0
_________________________________________________________________
dense_19 (Dense) (100, 128) 1605760
_________________________________________________________________
dense_20 (Dense) (100, 4) 516
=================================================================
Total params: 1,680,457
Trainable params: 1,680,457
Non-trainable params: 0
_________________________________________________________________
The probabilistic model involves only two random variables; latent variable \(\mathbf{z}\) and observation \(\mathbf{x}\). We put a Normal prior on \(\mathbf{z}\), decode the variational parameters of \(q(\mathbf{z}|\mathbf{x})\) and define the likelihood of the observation \(\mathbf{x}\).
In [44]:
with pm.Model() as model:
# Hidden variables
zs = pm.Normal('zs', mu=0, sd=1, shape=(minibatch_size, dim_hidden), dtype='float32', total_size=len(data))
# Decoder and its parameters
dec = Decoder(zs, net=cnn_dec)
# Observation model
xs_ = pm.Normal('xs_', mu=dec.out, sd=0.1, observed=xs_t, dtype='float32', total_size=len(data))
In the above definition of the generative model, we do not know how the
decoded variational parameters are passed to
\(q(\mathbf{z}|\mathbf{x})\). To do this, we will set the argument
local_RVs in the ADVI function of PyMC3.
In [45]:
local_RVs = OrderedDict({zs: (enc.means, enc.rhos)})
This argument is a OrderedDict whose keys are random variables to
which the decoded variational parameters are set, zs in this model.
Each value of the dictionary contains two theano expressions
representing variational mean (enc.means) and rhos (enc.rhos). A
scaling constant (len(data) / float(minibatch_size)) is set
automaticaly (as we specified it in the model saying what’s the
total_size) to compensate for the size of mini-batches of the
corresponding log probability terms in the evidence lower bound (ELBO),
the objective of the variational inference.
The scaling constant for the observed random variables is set in the same way.
We can also check the architecture of the decoding network as for the encoding network.
In [46]:
dec.model.summary()
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_11 (InputLayer) (100, 2) 0
_________________________________________________________________
dense_21 (Dense) (100, 2) 6
_________________________________________________________________
dense_22 (Dense) (100, 12544) 37632
_________________________________________________________________
reshape_7 (Reshape) (100, 64, 14, 14) 0
_________________________________________________________________
conv2d_transpose_19 (Conv2DT (100, 64, 14, 14) 36928
_________________________________________________________________
conv2d_transpose_20 (Conv2DT (100, 64, 14, 14) 36928
_________________________________________________________________
conv2d_transpose_21 (Conv2DT (100, 64, 28, 28) 16448
_________________________________________________________________
conv2d_23 (Conv2D) (100, 1, 28, 28) 257
=================================================================
Total params: 128,199
Trainable params: 128,199
Non-trainable params: 0
_________________________________________________________________
Inference¶
Let us execute ADVI in PyMC3.
In [47]:
# In memory Minibatches for better speed
xs_t_minibatch = pm.Minibatch(data, minibatch_size)
with model:
approx = pm.fit(
15000,
local_rv=local_RVs,
more_obj_params=enc.params + dec.params,
obj_optimizer=pm.rmsprop(learning_rate=0.001),
more_replacements={xs_t:xs_t_minibatch},
)
Average Loss = 2.2294e+07: 100%|██████████| 15000/15000 [23:47<00:00, 10.52it/s]
Finished [100%]: Average Loss = 2.2264e+07
I’ve checked the plot, seemed like convergence is not achived
Results¶
ADVI instance has the trace of negative ELBO during inference (optimization). We can see the convergence of the inference.
In [49]:
plt.plot(approx.hist);
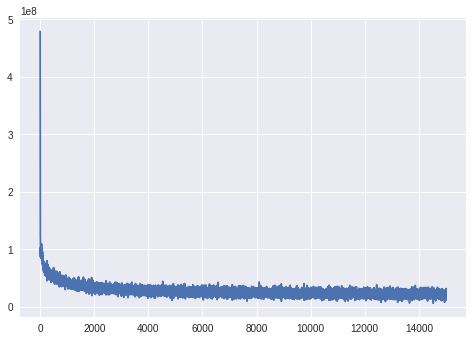
Finally, we see the distribution of the images in the latent space. To do this, we make 2-dimensional points in a grid and feed them into the decoding network. The mean of \(p(\mathbf{x}|\mathbf{z})\) is the image corresponding to the samples on the grid.
In [50]:
nn = 10
zs = np.array([(z1, z2)
for z1 in np.linspace(-2, 2, nn)
for z2 in np.linspace(-2, 2, nn)]).astype('float32')
xs = dec.decode(zs)[:, 0, :, :]
xs = np.bmat([[xs[i + j * nn] for i in range(nn)] for j in range(nn)])
matplotlib.rc('axes', **{'grid': False})
plt.figure(figsize=(10, 10))
plt.imshow(xs, interpolation='none', cmap='gray')
plt.show()
